Mutual information
The mutual information of two alignment columns is a measure of correlated mutations. We compute the mutual information of a pair of columns according to (Schneider 1986). This includes a correction term to account for the sampling noise for small number of sequences. The information R of a set of characters (here a column in the alignment) is the decrease in uncertainty H after reading that set of characters. The mutual information of two alignment columns i and j is the information from that column pair taken together minus the information of the alignment columns taken separately. The formula used is:
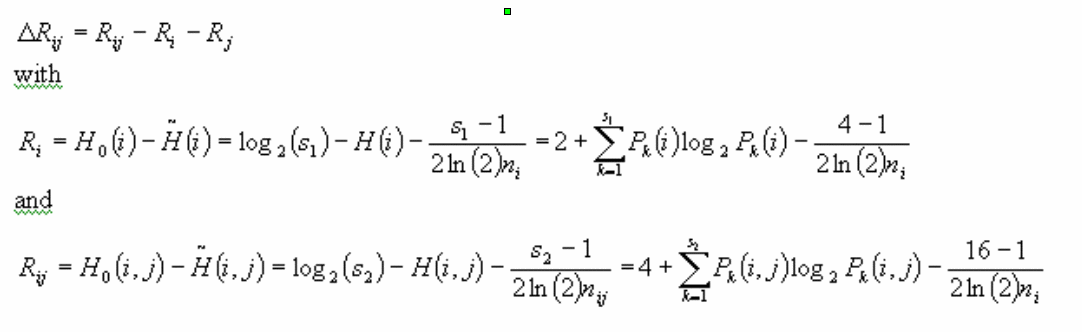
The alphabet sizes are s1=4 (4 letter alphabet ACGU) and s2=16 (sixteen possible base-pairs). The number of sequences which have a non-gap character at positions i and j is called nij (ni for only one column). Pk(i) denotes the probability of finding a certain characters of type k at position i; Pk(i,j) is the probability of finding one of the sixteen possible character pairs k at positions i and j. The probabilities are approximated by frequencies: (Pk(i): number of found characters of type k divided by the total number of non-gap characters in column i). H0 is the initial uncertainty of one column (H0(i)) or two columns (H0(i,j)). The initial uncertainty is approximated here as 2 bits for one column and 4 bits for two columns corresponding to a 4-letter and a 16-letter alphabet.
References:
Schneider T.D., Stormo G.D., Gold L, Ehrenfeucht A.: Information content of binding sites on nucleotide sequences. J Mol Biol. 1986. Apr 5;188(3):415-31.
Schneider T.D. and Stephens R.M. : Features of spliceosome evolution and functioninferred from an analysis of the information at human splice sites. J Mol Biol. 1992 (228):1124-1136.