Frequently Asked Questions
What is an RNA junction?
For the purposes of this database we have considered internal loops, bulges, kissing loops, and junctions to the 9th order (9-way.) Fig. 1 shows one of the 2,337 three-way junctions (3WJ) identified by JunctionScanner. A junction or branchpoint in RNA is defined, by the International Union of Biochemistry and Molecular Biology, as per Lilley et al. 1995, as "the point of connection between a number of different helical segments."
Fig. 1 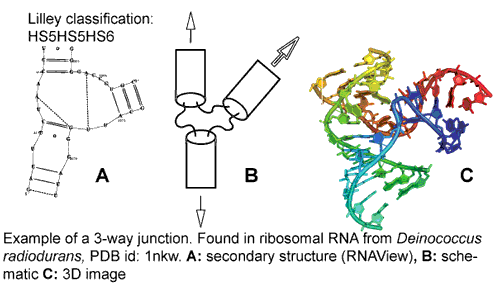
What are kissing loops and bulges/internal loops?
A kissing loop is an interaction (base-pairing) between two single-stranded portions of bases that pair with each other instead of the opposite strands in their respective helices. An internal loop is a small segment of unpaired bases in a longer paired helix. Bulges are regions in which a strand of a helix has extra inserted bases with no free nucleotides in the opposite strand.
How many junctions have been found?
Our group has documented and made available 13127 junction and kissing-loop structures.
How can I link to a particular junction?
https://rnastructure.cancer.gov/rnajunction/JunctionPage.php?jid=n
n is the junction id which corresponds to the junction you are looking for.
Who created RNAJunction?
The web site and database were implemented by Bob Hayes, a student in Bruce Shapiro's group, RNA Biology Lab, National Cancer Institute, NIH. The junction data was extracted from the available RNA structures in the PDB database by Eckart Bindewald using a program he developed for that purpose. Energy minimizations on all the junctions were performed by Wojciech K. Kasprzak. Both raw and energy minimized junction files (PDB) are available for downloading.